
- VMD TUTORIALS HOW TO
- VMD TUTORIALS MANUAL
- VMD TUTORIALS SOFTWARE
- VMD TUTORIALS CODE
- VMD TUTORIALS FREE
Color by ID (red), Select again drawing method Isosurface with an Isovalue of -3. * Create a new representation, Color by ID (blue), Select drawing method Isosurface. * In Graphics Representation, change the standard representation to New Cartoon. Select the corresponding dx grid file from your apbs output Press Load. In the respective pqr files, what are the net charges for the two proteins _?_ * To visualize the electrostatic potential, select in the vmd main window the molecule 1yer. * Repeat for both structures and unpack the *.dx.gz file. You might want to look at the program output and error logs if there are errors or warnings. * Again, save the output files, espcially the *.dx.gz grid files. * Save the output files (xxxx_aligned.in, xxxx_aligned.propka and xxxx_aligned.pqr) on disk.

* Upload one of the aligned coordinate files.Ĭhoose the force field (AMBER) and naming scheme (AMBER), and click ’Submit’. The proteins should show up as aligned entities. Direct questions to resource is provided by the Center for Theoretical Biological Physics.Goal: Visualize possible interaction sites on proteins * Start VMD again and use the aligned Proteins from the previous section (load the previously saved coordinates).
VMD TUTORIALS HOW TO
A tutorial explaining how to build lipid bilayers as well as insert membrane-bound proteins is available on the VMD web site. As well, we assume you are a scientist and read the appropriate literature.
VMD TUTORIALS MANUAL
We encourage users to refer to the Amber Reference Manual for syntax and detailed explanations.
VMD TUTORIALS SOFTWARE
Documentation and examples are included in the download package. A plugin is available for the Visual Molecular Dynamics (VMD) program that can currently build POPC and POPE bilayer systems. Amber Tutorials Below are tutorials prepared by the Amber developers to help you learn how to use the Amber software suite.
VMD TUTORIALS FREE
Features include, but are not limited to, free energy in multiple dimensions, umbrella bias removal and free energy perturbation, ensemble average of a user-defined quantity, customization of data input methods. PyWham (2/10/15) Another implementation of the Weighted Histogram Analysis Method written in Python. Found in either of the SMOG enhanced Gromacs versions:Ĥ.5.3 or the 5.0.4. G_kuh - calculates Q, the fraction of native contacts for a trajectory (.xtc).
VMD TUTORIALS CODE
Examples and source code can be found in the SMOG 2 distribution. Functionality is described in the SMOG 2 Manual ( PDF). Includes options to remove harmonic umbrella biases. WHAM (version 1.) An implementation of weighted histogram analysis that is tailored for use with SMOG to compute thermodynamic quantities. Also, please read the comments inside of the script to make sure you use it properly. Always double check the value of CUTOFF before using this script. This may not be the definition you want to use. **CAUTION** This script allows you to define contacts as any native pairs within CUTOFF times the native distance. Also, please read the comments inside of the script to make sure you use it properly.Ĭontacts.CA.pl (11/02/09) determines the number of native contacts formed as a function of time for your trajectory for an C-alpha SBM (i.e. Contributed tools are always appreciated.Ĭontacts.AA.pl (10/26/08) determines the number of native contacts formed as a function of time for your trajectory for an All-atom SBM (i.e. Many produce text files formatted for use with Grace (a very powerful 2-D plotting tool).Īdditional scripts/programs: Scripts for tasks not easily available through Gromacs and VMD will be posted here over time. Gromacs also includes a large number of analysis programs with their distribution. This will load the xtc file into the gro structure.Ĭalculating quantities from your simulations: If you prefer to load the structures from the command line, issue "vmd GRO_o XTC_FILE.xtc".
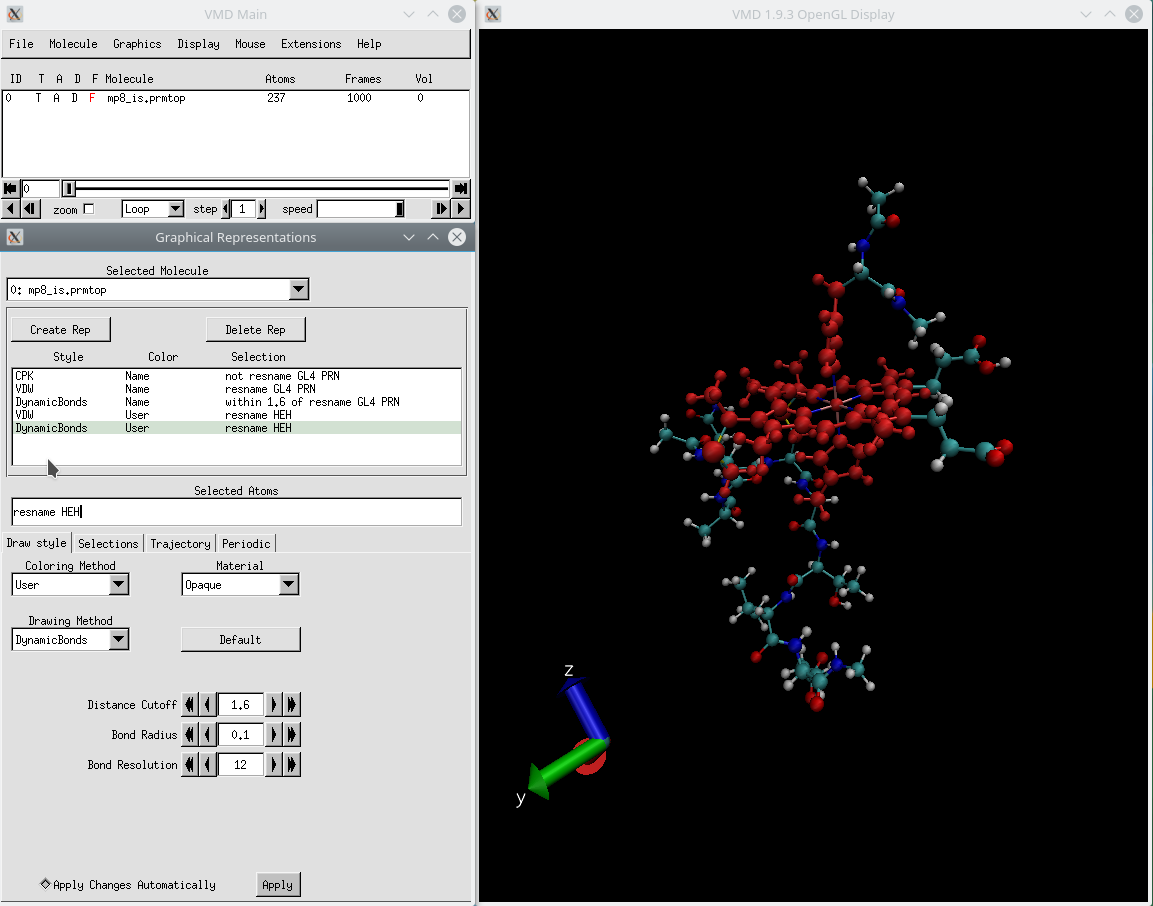
gro (coordinate) file and then select "load data into molecule" and load the. To watch a Gromacs trajectory in VMD, simply load the. It also provides a variety of easy to use GUI tools for trajectory analysis.
Visualizing your trajectories: VMD is our favorite software for molecular dynamics visualization.


 0 kommentar(er)
0 kommentar(er)
